Migration from Bacterial and Plant&Fungal CSDB
 +
+
 →
→

|
The persistent record IDs of the ancestor databases have remained intact upon migration to the merged database, so you do not need to update your references to records in BCSDB or PFCSDB.
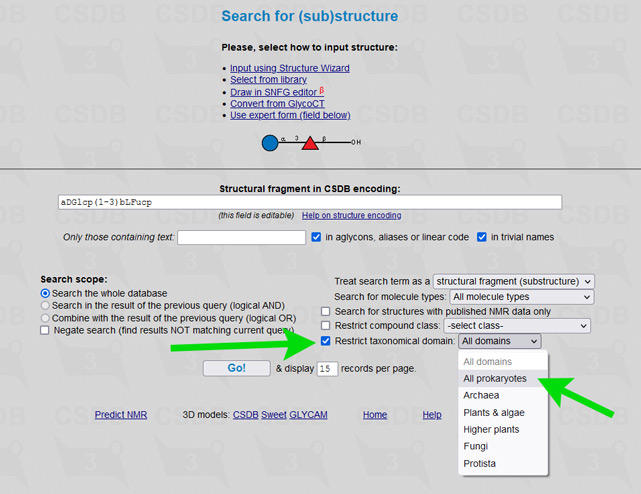
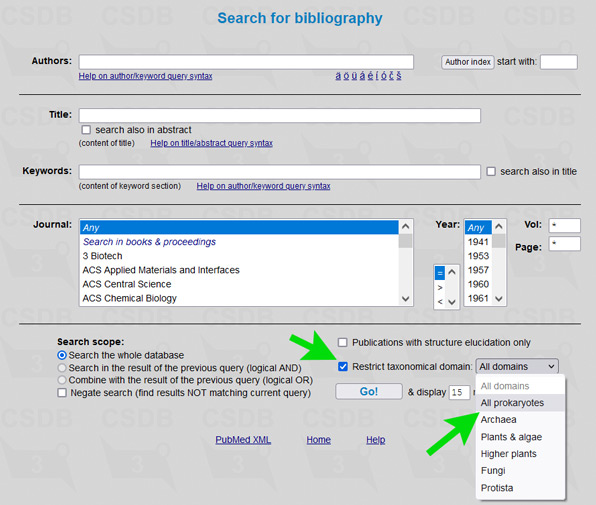
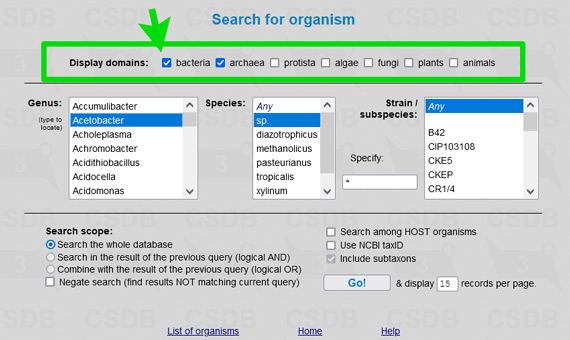
The user interface of the merged database is the same as of its ancestors, including all its tools and add-ons. To simplify restriction of search results to a certain part of a tree of life, an element Restrict domain has been added to search forms (see → green arrows on screenshots below).
For example, to limit results of a substructure search to bacterial and archaeal structures only (as it was in Bacterial CSDB), check the checkbox Restrict taxonomical domain and select All prokaryotes from the dropdown list. In composition and bibliography search, operate similarly. In taxonomy search, you have to select a genus first (as it was in BCSDB); to reduce the list of genera to a certain taxonomic group, check or uncheck the checkboxes in the Display domains group.
Home