CSDB usage: glycosyltransferases
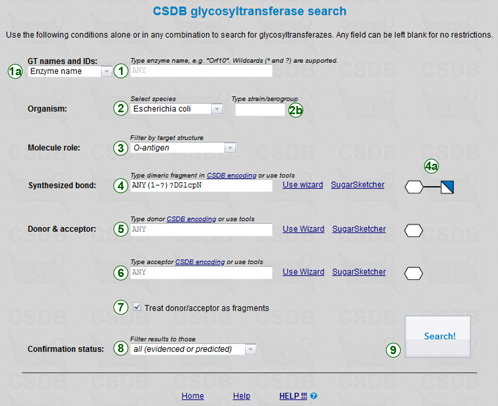 |
The glycosyltransferaze search form (←) provides several search criteria:
Names / IDs (1): enzyme name or enzyme group name, CAZY family, enzyme Uniprot ID, gene GenBank ID, internal CSDB GT identifier. The user can select only one of these criteria (1a), as they are never expected to intersect. Wildcards (* for any number of any characters and ? for strictly one character) can be used in the names. Ranges and lists are supported for internal GT IDs, e.g. 1-10,20.
Organism: you can select the origin species (2) and optionally type subspecies/strain (2a). Currently, only E. coli and A. thaliana are available, however more data will be returned if species is set to ANY.
Molecule role: allows filtering enzymes to certain cellular roles of a product they synthesize, or its analog. This selector (3) contains roles like O-antigen, lipid A, CPS, etc.
Synthesized bond: the main result of glycosyltrabsferase activity presened as a dimeric fragment with residues linked by a specific bond. It can be typed directly in CSDB Linear notation (4), or entered using the structure wizard (a dedicated glycan structure input tool), or drawn in Sugar Sketcher (an SNFG glycan structure editor). The corresponding dimeric fragment in the SNFG format is previewed on the right (4a).
Donor (5) and/or acceptor (6): the structure of the carrier of the transferred monosaccharide, and of the substrate it is carried to. The input and preview options are the same as for the synthesized dimeric fragment. Checkbox Treat as fragments (7) determines whether to treat the donor and acceptor as structural fragments (default) or as whole structures. Please note that the exact donor and substrate information is not always known, so strict interpretation of the structures achieved by unchecking this option can significantly limit the output.
Confirmation status (8): the type of evidence for the glycosyltransferase activity (see below). Possible filters include: Confirmed or predicted - no restrictions on the methods of investigation; Confirmed experimentally - GTs with activity confirmed by any experimental methods; With direct or indirect evidence - GTs with activity supported by direct, semi-direct, or indirect evidences; Confirmed strictly in vivo - GTs with activity supported by direct evidences.
Pressing the button (9) runs the query. If multiple criteria are specified, the results are intersected using boolean AND.
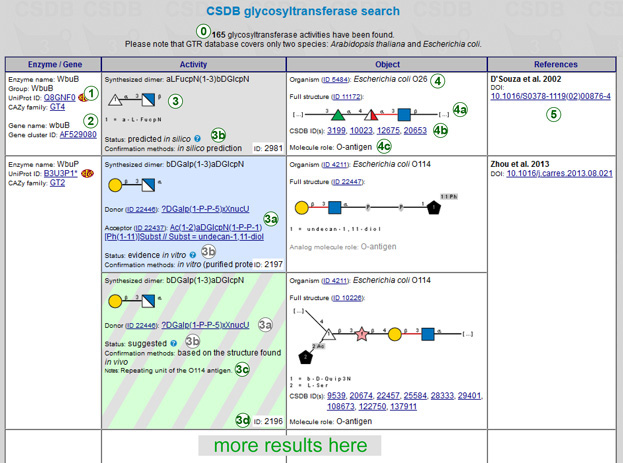 |
The number of found enzymatic activities (0) is reported in the header. The resulting table (→) contains the following data:
Enzyme / Gene (column 1): available data on the enzyme (1), such as name, group name, and links to the UniProt entry and to CAZY family, and on the gene (2), such as name, and links to the GenBank entry or to the Genbank cluster. Clicking on the ID opens the corresponding external database entry. Every enzyme-gene pair has at least one strict reference to other databases. Click  beside a Uniprot ID or NCBI Genbank gene ID to open a gene and its orthologs in KEGG genes database.
beside a Uniprot ID or NCBI Genbank gene ID to open a gene and its orthologs in KEGG genes database.
Activity (column 2): the main answer produced by CSDB glycosyltransferase module. Every activity has a unique GT ID displayed in the bottom right corner of the cell (3d). These IDs are persistent and can be used to reference a certain entry in CSDB_GT. The synthesized bond (dimer) is displayed in CSDB Linear and SNFG formats (3). Please note that, according to the CSDB ideology, monovalent substituents, such as residues of acetic acid, are always treated as separate residues, e.g. Ac(1-2)bDGlcpN is a dimer.
Donor and acceptor information (3a) is presented in the form of full or partial structures, if available. IDs are linked to compound pages in CSDB, while structures in the CSDB Linear notation are linked to their graphic display.
The confirmation status (3b) reflects the trustworthiness level of the activity determination. The cell background is color-coded according to this value, which can be the following:
- predicted in silico - predicted with bioinformatic means
- evidence in vitro - evidenced by an in vitro reaction of a pure enzyme with a substrate OR by reaction in the lysate of cells overexpressing the enzyme (details are given in confirmation methods)
- indirect evidence in vivo - evidenced in the source organism by indirect methods, such as mutagenesis and overexpression
- semi-direct evidence in vivo - evidenced in vitro with substrates, which differ from those naturally occuring in the organism, AND in the source organism by indirect methods, such as mutagenesis and overexpression
- direct evidence in vivo - evidenced in vitro, AND in the source organism by indirect methods, such as mutagenesis and overexpression
- suggested - an enzyme is present in the organism, AND its activity is suggested on the basis of carbohydrate structures found in the organism in vivo
Experimental methods used to confirm the activity, and additional notes (3c) are provided if available.
Object (column 3): includes the biological reference and the structure in the synthesis of which the activity is implicated. Species and subtaxa (4) are linked to CSDB organism pages. Organs or tissues are displayed where available.
The full structure or its characteristic fragment is visualized graphically and has a link to the corresponding CSDB compound ID (4a).
If the structure is present in CSDB as real (i.e. was deposited independently from CSDB_GT, in contrast to virtual structures that do not have a bibliographic reference), one or more corresponding CSDB ID links are displayed to allow retrieval of all data related to this structure in this organism.
Molecule role (4c) of the synthesized object or its analog is also provided, if it could be derived from the publication on a GT activity.
 |
Reference (column 4): imprints of original publications, where the activity was shown (5). The most relevant reference is marked as main and displayed in bold. Every reference has at least one identifier in the external bibliographic databases (DOI, Pubmed ID or Internet address).
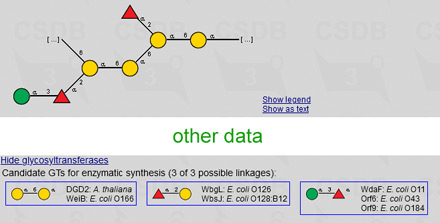
When a structure is displayed within a record, or as a result of structure or composition search, it has a link Show glycosyltransferases to open a preview (←) of candidate glycosyltransferases that can be used in the enzymatic sythesis of this structure. The preview lists source organisms and names of enzymes grouped by activity. More data are available by clicking on the SNFG image of a synthesized bond. Absence of the link Show glycosyltransferases means that there were no matching GTs found.
Simplest API and export
To address certain records, please use the following URL: http://csdb.glycoscience.ru/database/core/search_gtr.php?gtr_ids={id list}, where {id list} is a comma-separated list of GTR IDs or their ranges, e.g. 1000-1004,2010,3012).
To search for glycosyltransferazes that can be potentially used to synthesize bonds in a certain structure, please use a GET parameter cid to specify a CSDB compound ID of a structure of interest, or POST parameter full_structure to specify a structure itself (in CSDB Linear notation).
To export the data from the search result table, use a link Export TSV in the page bottom. It opens the container with tab-separated data for the displayed records, and selects the text. You can copy (Ctrl-C) the data to clibpoard and paste it to an external spreadsheet software for further processing. The first row explains column names. To extract the data in the automated manner, look for DIV with ID='tsv' in the server response.
Home