| Enzyme / Gene |
Activity |
Object |
References |
|---|
Enzyme name: WaaB
UniProt ID: P27127 
CAZy family: GT4
Gene name: waaB / rfaB
Gene GenBank ID: 948144*  | Synthesized dimer: DGalp(1-6)DGlcp
?DGlcp)
Status: indirect evidence in vivo
Confirmation methods: mutation (deletion)
ID: 2107 Notes: WaaB participates in the synthesis of glycoform I of the LPS core.
| Organism (ID 23): Escherichia coli K12
Full structure (ID 34422):
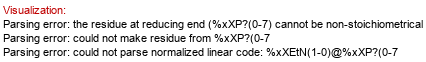aXKdop(2-4)[aXLDmanHepp(1-6)?DGlcp(1-2)?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%25xXEtN(1-%25P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A&highlight=5~6~?DGal)
Molecule role: core | Klein et al. 2011
DOI: 10.1074/jbc.M111.291799
|
Synthesized dimer: DGalp(1-6)DGlcp
?DGlcp)
Status: indirect evidence in vivo
Confirmation methods: mutation (deletion)
ID: 2108 Notes: WaaB participates in the synthesis of glycoform IV of the LPS core.
| Organism (ID 23): Escherichia coli K12
Full structure (ID 34423):
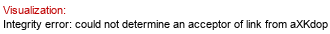[??Rhap(1-5)]aXKdop(2-4)[?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%25xXEtN(1-%25P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A&highlight=5~6~?DGal)
Molecule role: core |
Synthesized dimer: DGalp(1-6)DGlcp
?DGlcp)
Status: indirect evidence in vivo
Confirmation methods: mutation (deletion)
ID: 2109 Notes: WaaB participates in the synthesis of glycoform V of the LPS core.
| Organism (ID 23): Escherichia coli K12
Full structure (ID 34424):
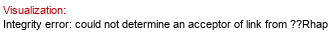aXKdop(2-4)[%25xXEtN(1-%25P-7)]aXKdop(2-4)[?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%25xXEtN(1-%25P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A&highlight=5~6~?DGal)
Molecule role: core |
#GTR ID Enzyme name Enzyme group Enzyme Genbank ID Derived ID mark Enzyme Uniprot ID Derived ID mark CAZY family Gene name Gene Genbank ID Derived ID mark Gene cluster Full structure CSDB structure ID Side product CSDB structure ID Full structure Full structure is fragment (0|1) Synthesized bond Synthesized bond location (from root ~ subst position) Donor structure ID Substrate structure Confirmation status Methods used Notes Molecule role Molecule is a model of role (0|1) Molecule is a precursor of role (0|1) Main CSDB record ID Additional CSDB record IDs CSDB organism IDs Organism names Organs and tissues References (;-separated)
2107 WaaB P27127 GT4 waaB / rfaB 948144 * 34422 %xXEtN(1-%P-7)aXKdop(2-4)[aXLDmanHepp(1-6)?DGlcp(1-2)?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%xXEtN(1-%P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A 0 ?DGalp(1-6)?DGlcp 5~6 indirect mutation (deletion) WaaB participates in the synthesis of glycoform I of the LPS core. core 0 0 23 Escherichia coli K12 Klein et al. 2011 (doi: 10.1074/jbc.M111.291799)
2108 WaaB P27127 GT4 waaB / rfaB 948144 * 34423 aXKdop(2-4)[??Rhap(1-5)]aXKdop(2-4)[?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%xXEtN(1-%P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A 0 ?DGalp(1-6)?DGlcp 5~6 indirect mutation (deletion) WaaB participates in the synthesis of glycoform IV of the LPS core. core 0 0 23 Escherichia coli K12 Klein et al. 2011 (doi: 10.1074/jbc.M111.291799)
2109 WaaB P27127 GT4 waaB / rfaB 948144 * 34424 ??Rhap(1-5)aXKdop(2-4)[%xXEtN(1-%P-7)]aXKdop(2-4)[?DGlcp(1-3)[?DGalp(1-6)]?DGlcp(1-3)[aXLDmanHepp(1-7),P-4)]aXLDmanHepp(1-3)[%xXEtN(1-%P-4)]aXLDmanHepp(1-5)]aXKdo(2-?)Subst // Subst = lipid A 0 ?DGalp(1-6)?DGlcp 5~6 indirect mutation (deletion) WaaB participates in the synthesis of glycoform V of the LPS core. core 0 0 23 Escherichia coli K12 Klein et al. 2011 (doi: 10.1074/jbc.M111.291799)

