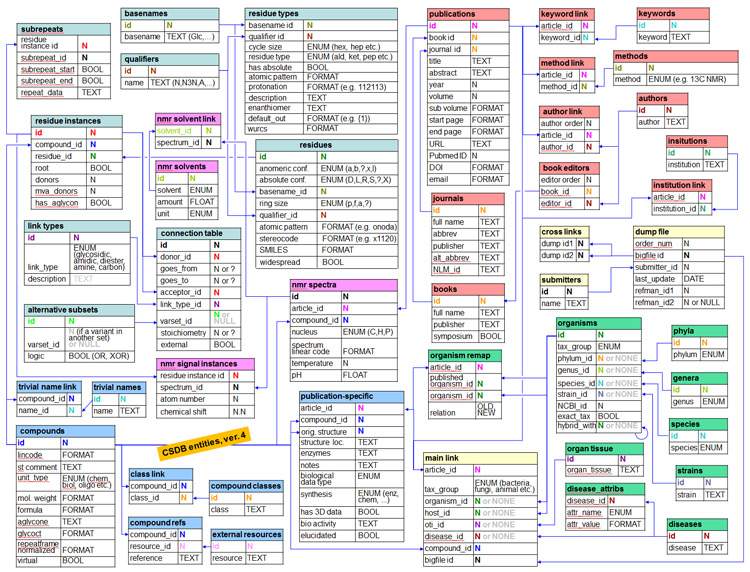
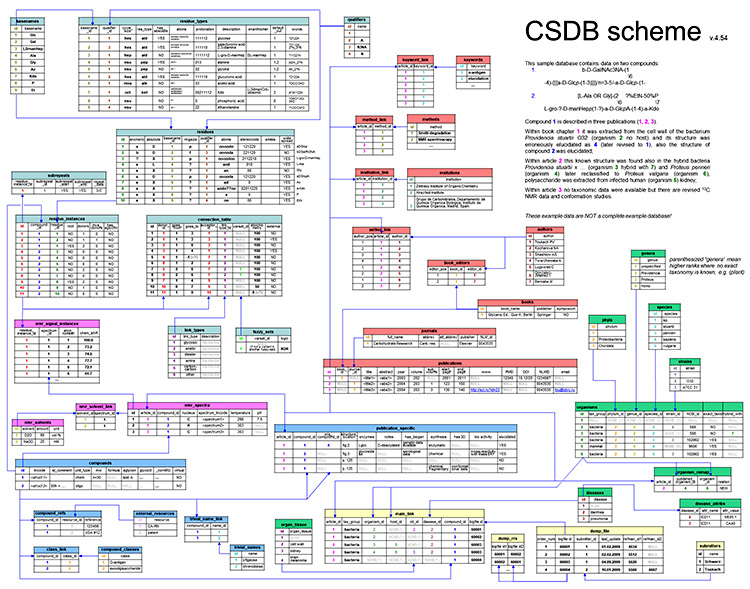
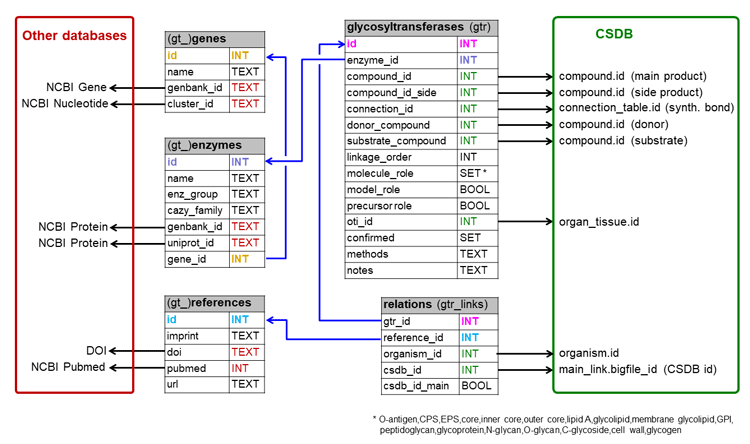
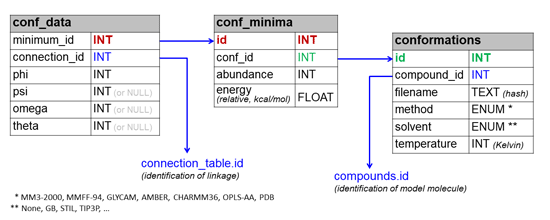
This section describes the architecture of CSDB, data formats and other technical details. For user help, please refer to Basic usage and Advanced features.
CSDB utilizes a MySQL relational database to store data. The connection table approach is used for structures.
Click on the core entity relationship scheme to enlarge it (PDF).
The above figure depicts relationaships between data from scientific publications and their indices. NMR-spectroscopic and structural data derived by statistical processing, theoretical prediction, and format conversion are gathered in a separate set of tables not presented in the figure. Background of table headers stands for data category:
The example database is explained (entities + data) in the figure (click to enlarge to A3 PDF):
Glycosyltransferase module is driven by a separate set of tables interlinked with CSDB and other databases:
Conformation map module is driven by a separate set of tables interlinked with CSDB:
CSDB contains four special tables for caching residue connection topologies (unlisted on the scheme). To view pre-generated topologies, click here.
The carbohydrate structure can be published explicitly (as a figure, scheme, IUPAC name) or implicitly (using a trivial name, free text or even only implied by authors). The presence of a structure in a publication can be expressed as: elucidation of primary structure or conformation; suggesting a structural motif; other studies, including biological activity; synthesis or modeling of a structure; assignment of structure to another taxon; referencing or reviewing a structure in regard to its biological role or other properties.
The CSDB dump is a backup of the database and a reference file for all the database content. It is a human-readable UTF-8 text file; content corrections and error-tracking are usually performed on this file. The dump file can contain one or more records (including a complete dump that contains all CSDB records) separated by two blank lines. Lines starting with # symbol are comments for annotators and are not processed.
Every record is a unique combination of a molecular structure (ST* + AG fields) and a paper in which this structure is dicsussed. Each record represented by several lines, one line per field. The line starts with the field name followed by colon (:) and field content. Line breaks inside the field are not allowed. Below is the explanation of fields. Please refer to the data submission page and to the example record at the end of this page for more details on the meaning of fields. Fields that cannot be empty are in bold.
| Field | Explanation |
|---|
| ID: |
Unique permanent CSDB record ID. Records that have some unresolved problems are marked by star (*; they are not processed until corrected). Records associated with publications that contain no information on structure of natural carbohydrates should be marked with two stars and an explanation why this record is excluded from the database (e.g. ID: 100 ** structure not found; such records are not processed at all).
|
| TH: |
1 if the structure was elucidated or revised in the publication (even by poor methods),
0 if the structure discussed in the publication was elucidated elsewhere (e.g. this publication is a review or biological study of a known structure).
If a known structure was identified but no elucidation details are provided, use TH=0
If a known structure was confirmed/revised by described elucidation, use TH=1
If a structure was revised but no elucidation details are provided, and the revised structure is already known, use TH=0
If a structure is supposed or arbitrary but authors obtained the structural data and publish the structure in the paper, use TH=1 and ST2=MOTIF.
If only a fragment was elucidated, while a bigger structure is known or supposed from elsewhere, use TH=1 and ST2=FRAGMENT for the fragment itself, and TH=0 for a bigger structure.
If the structure was elucidated erroneously, use TH=1, ST1=proper structure, ST1ORIG=erroneous structure. In this case, please specify in NT, how the structure was revised (basing on the NMR data, revised later from [ref...], etc.)
Example:
Statements in the paper:
A new compound 1 was extracted from fungus X, and its structure was elucidated by NMR. Moreover, known compounds 2-6 were extracted from the same fungus. Compound 2 was shown to have a certain sequence (6-mannosylated and linear glucose residues interleave one-by-one; it was previously reported only as a structural motif of 50%-6-mannosylated glucan). In compound 3, a previously unknown position of a methoxy group in the aglycon was determined. Structure of compound 4 was revised, and differs from the previously reported one by presence of a rhamnose instead of mannose in the side chain. Compound 5 was found identical to a known glycoside from plant Y. Compound 6 was earlier synthesized chemically but now it was found in fungus X. From our studies, compound 7 was identified as 3-rhamnosylquercetin (elucidated elsewhere), but not 1-rhamnosylquercetin as reported earlier. Structural motif (but not exact sequence) of compound 8 was elucidated by NMR as a shizophyllan (known class of compounds).
Annotation:
TH:1 for compounds 1-4;
TH:0 for compounds 5-8.
|
| AU: |
Comma-separated list of authors. Last name goes first, then initials without dots. National characters are supported (e.g. Müller AB).
|
| TI: |
Title of publication (article, chapter, book, symposium thesis). Please do not capitalize every word. If a title was copy-pasted from PDF, check it for missing spaces and merged characters.
|
| JN: |
Journal name without abbreviations. If unsure, please use CSDB journal list to check the spelling.
For book publications, use the following format: JN: BOOK: book name (series: series name, if known; otherwise omit these parentheses); Eds.: comma-separated editors; Publisher.
For symposium thesis, use the following format: JN: SYMP: symposium name (symposium number : year : place); Eds.: comma-separated editors; Publisher.
If editor/publisher is not known, type semicolons only, e.g. SYMP: Eurocarb (17th : 2013: Tel-Aviv);;
|
| PY: |
Publication year. If paper publication year differs from ePub year, use the former one.
|
| VL: |
Volume number. If the volume imprint contains a subvolume/issue, e.g. 36(5), specify the subvolume/issue in parentheses.
For books, specify the volume number outside parentheses and the chapter number inside parentheses. The book series number should be included in the book title (JN).
|
| PG: |
Hyphen-separated start and end page numbers. If the end page is unknown, use the start page only.
For imprints with article number instead of page number, use ID keyword, e.g. ID 35
|
| RL: |
References to bibliography: comma-separated list of resource-identifier pairs (resource identifier is before colon, reference is after colon, e.g. PMID:123456789). Allowed resources are: PMID (NCBI PubMed ID), DOI (DOI code), URL (www address), NLMID (NCBI NLM ID of book or chapter).
|
| EA: |
E-mail of the corresponding author, e.g. address@gmail.com; My Name <my_email@my_server.ru>
|
| AD: |
Semicolon-separated list of author affiliations (institution, city, country). Each affiliation should be listed only once, the order is arbitrary.
|
| AB: |
Publication abstract. Please change line breaks to blank spaces. National characters are supported. If abtract was copy-pasted from PDF, check it for missing spaces, merged characters and extra line breaks. All chemical structures should be either specified in linear form or replaced with the word /structure/.
|
| ST1: |
Chemical structure in CSDB linear encoding (see Structure encoding rules for details).
If you used Subst for a fully defined substructure, except simplest cases, it is strongly recommended to add a SMILES code after the second equation mark, e.g. aDRibf(1-3)Subst // Subst = questin = SMILES CC1=CC(=C2C(=C1)C(=O)C3=C{3}C(=CC(=C3C2=O)OC)O)O. Please note that SMILES should describe a complete residue, including hydroxy or other group that is removed upon formation of the bond to carbohydrate moiety. Please indicate the atoms that form bonds with other residues by figure brackets and atom number ({3} in this example) before the corresponding carbons. This is needed to unambiguify authors' atom enumeration.
|
| (optional) ST1ORIG: |
Originally published erroneous chemical structure in CSDB linear encoding (the field is present only if ST1 contains the revised structure).
In case the error is in the aglycon, specified by its SMILES code, omit aglycon name from explanation (e.g. Subst = SMILES CC{2}C(N)C for wrong struicture in ST1ORIG, but Subst = 2-butanol = CC{2}C(O)C for correct structure in ST1)
|
| ST2: |
Structure type: OLIGO (oligomeric structure), CHEM (chemical repeating unit of a polymer), BIOL (proven biological repeating unit), SBIOL (suggested biological repeating unit), MONO (oligomeric structure with a single carbohydrate entity), HOMO (repeating unit of a homopolymer), CYCLO (repeating unit of a cyclic polymer), FRAGMENT (poly- or oligomeric fragment of a bigger structure), MOTIF (supposed, idealized or arbitrary structure; exemplary structure with certain pattern of side chains, where multiple interpretations are possible; exemplary structure with explicit values of n/m/k/etc indicating the polymericity of subfragments - in this case add a comment, e.g. NT: motif of polymeric structure 29 when n=m=1, see ID #####).
Biological repeating unit can be suggested basing on the same structure from the same organism in some other record (BIOL) or on general knowledge, e.g. if a repeating unit contains a single aminosugar, its biological repeat is likely to start with this aminosugar. In E. coli, presence of genes GNE or GNU indicate biological repeat start with GalNAc.
|
| ST3: |
For polymers: Polymerization degree preceded by n=, e.g. n=12-15, or molecular mass in Daltons. Ranges and relations are supported. e.g. 10000-30000 or >30000.
For oligomers: Molecular mass with or without ion type in square brackets, e.g. 9813 [M+H]+.
Separate multiple values by comma.
|
| SL: |
Structure location inside the publication, e.g. structure 1, HPLC fraction 2, compound 7a, fig. 3, p.456, etc. Except structure itself, please indicate tables with the NMR assignment.
|
| AG: |
Aglycon information (what is attached to the reducing end and by which position, if known), e.g. (->6') lipid A or inner core, bDGalpN C6. If the aglycon is a single residue present in the monomeric namespace (e.g. Allyl) or more than one residue is attached to the aglycon, encode it in the structure (ST1). AG field is proposed for describing aglycons if aglycon structure could not be fully determined, or when aglycon caps the reducing end of a polymeric glycan. In all other cases it is advised to encode aglycon as Subst alias, and add its SMILES code in the explanation (see example above in ST1).
Leading parentheses indicate the attachment site in the aglycon (e.g. (->3) sapogenin F1). Greek letters, single and double quotes are allowed. |
| MF: |
Molecular formula (for mono- and oligosaccharides only). Carbons first, then hydrogens, then alphabetically.
|
| NMRH: |
1H NMR assignment data (see NMR data storage for details). Paste structure into NMR template generator to generate a template to fill with chemical shifts. If the spectrum was published but chemical shifts were not picked, , type present in publication.
Proton enumeration within residues follows carbon enumeration. If a certain position does not have protons, use a hyphen (-) at corresponding position in a proton spectrum. If a carbon has non-exchangeable protons but proton chemical shift could not be observed, use a question mark (?). Chemical shifts of two protons attached to the same carbon are separated by hyphen in the ascending order. Protons attached to heteroatoms should be recorded only if this heteroatom has a number in a carbon sequence, e.g. two parts of a carbon skeleton are connected via NH group. Exchangeable protons (-OH, -COOH, -NH2, etc.) are always omitted.
The spectrum should correspond to the exact structure specified in ST1. Please avoid situations like a spectrum deposited for an oligomer, while structure is a polymer with the same repeating unit (because capping groups are different), or a spectrum deposited for a fragment, while structure contains other moieties attached, etc. If the structure contains non-stoichiometric moieties (residues denoted by %), provide spectra for the structure variant with these moieties attached.
|
| NMRC: |
13C NMR assignment data (see NMR data storage for details). Paste structure into NMR template generator to generate a template to fill with chemical shifts. If the spectrum is published but chemical shifts were not picked, type present in publication. For carbon enumeration, refer to Monomer namespace. If you provide a spectrum for a residue alias (Subst, etc.), ensure to explain the atom enumeration in NT (according to IUPAC; or according to the Fig. # in the paper; web-reference on Wikipedia etc.)
For unresolved chemical shifts, use a question mark (?) at corresponding position.
The spectrum should correspond to the exact structure specified in ST1. Please avoid situations like a spectrum deposited for an oligomer, while structure is a polymer with the same repeating unit (because capping groups are different), or a spectrum deposited for a fragment, while structure contains other moieties attached, etc. If the structure contains non-stoichiometric moieties (residues denoted by %), provide spectra for the structure variant with these moieties attached.
|
| NMRT: |
Temperature used to carry out NMR experiments, in Kelvins. If carbon and proton spectra were recorded at different temperature, separate values with comma and specify nuclei in parentheses, e.g. 313(H), 298(C).
|
| NMRS: |
Solvent used to carry out NMR experiments (chemical formula or abbreviation). Separate mixed solvents with slash (e.g. CD3OD / D2O), starting from those with greater part. If ratios are known, write them before the solvents: 90%D2O / 10%H2O / 25mM NaCl or vol 67%CDCl3 / vol 33%DMSO-d6, etc. If a reference standard is not TMS, specify it as another solvent: D2O / DSS. pH value can be appended after semicolon: D2O; pD 7.5. To view the canonical names of most used solvents, click on Coverage link at the NMR simulation page.
|
| SO: |
Semicolon-separated list of biological sources of the structure, without taxon abbreviations. The first word in every source is interpreted as genus, the second is for species, all the other words (third and subsequent ones) are combined subspecies, serogroup, and/or strain.
If a taxon was renamed or an organism was reclassified after the data were published, specify the newer name in curly brackets after the organism, e.g. Enterobacter sakazakii O6 {NEW: Cronobacter sakazakii O6}. Older synonyms are specified as {OLD: ....}. The published taxon name should be always outside curly brackets; any number of OLD and/or NEW terms can be combined inside curly brackets. Please note, that a renamed taxon has almost always the same rank as the previous name (don't rename strains to species etc.).
If species name is unknown, while strain is known, specify sp. for the species name. If only genus or genus/species information is available, incomplete taxonomic annotations are allowed. If only a rank higher than genus is known (e.g. family), parenthesize it. For hybrid organisms, combine two taxa with a star (A * B or e.g. A-new * B {OLD: A-old * B}).
In the third part of taxon, use the following order and abbreviations for space-separated subdivisions: subspecies (ssp.), serovar (sv.), pathovar (pv.), biovar (bv.), serogroup (value without prefix, e.g. O1), strain (value or space separated values without prefix), mutant (in parentheses, e.g. (ΔgalE mutant)). The strain can be collection identifier (collection name and strain ID, e.g. ATCC 123, preferrable) or recognized or authors' designation (e.g. ABC123 or Nagasaki).
Single taxon examples:
(bacteria) - only kingdom is known
Proteus - only genus is known
Proteus penneri - only genus and species are known
Proteus penneri O22 - genus, species, serogroup
Acinetobacter haemolyticus ATCC 19606 - genus, species, strain from ATCC collection
Escherichia coli O86:B7 ATCC 12701 - genus, species, serogroup, strain
Citrobacter frendii PCM1555 (ΔgalE mutant) - genus, species, mutant strain
Salmonella enterica ssp. enterica sv. Typhimurium TV119 - genus, species, subspecies, serovar, strain
Haemophilus parasuis sv. 5 Nagasaki - genus, species, serovar, named strain or other subdivision
Pseudomonas sp. WAK-1 - species is uknown, genus and strain are known
|
| KD: |
Before slash (/): Taxonomic domain (bacteria,protista,archaea,fungi,algae,plant,animal,mammal,human).
After slash (/): Taxonomic phylum.
If there are multiple organisms of different domains or phyla, separate values with comma (in this case number of values should correspond to the number of organisms, including remappings in curly brackets).
For hybrid organisms (A*B), use phylum for ancestor B (one on the right).
|
| OTI: |
Organ, tissue, secret, or other biomaterial from which the structure was extracted. For microorganisms, organella or cell part can be specified. Organs of host organisms are also allowed. If life state is specified (embryo, culture broth, promastigote etc.) specify it in this field preceeded by Life stage: keyword. Use only singular nouns, separate multiple entries with comma, and do not use commas and parentheses within terms.
|
| DSS: |
Disease of the host organism associated with the structure or its biological source, or disease of a patient from which the structure was extracted. Please separate multiple diseases with semicolon and provide attributes in square brackets when possible. Comma-separated list of attributes can include: ICD11: ICD-11 code, Life stage: life stage of the host organism, Sex: male or female. If no disease is known but an infectious agent has an ICD-11 code, specify a disease as infection due to <taxon> [ICD11: X<...>]. Please do not combine multiple diseases in one title (like "colitis and diarrhea"). Examples: cholera [ICD11: 1A00, ICD11: XN7N1]; neonatal aspergillosis [ICD11: KA63.1, ICD11: XN0WC, Life stage: neonatal]; infection due to Proteus penneri [ICD11: XN7PE, Sex: male]. Always use canonical disease names, as listed in CSDB disease list.
|
| HO: |
Systematic name (genus and species) of the host organism in which the microorganism (specified in SO) was found. If multiple, separate hosts with a semicolon (;).
|
| NC: |
(Trivial) name of the compound. Greek letters, apostrophs and quotes are supported. Separate multiple names by semicolon.
|
| CC: |
Comma-separated list of classes and roles of the compound, e.g. O-antigen, EPS, phosphoglycolipid, GPI-anchor etc. For spelling and word order of popular classes, please refer to Class abundancy table
|
| MT: |
Comma-separated list of methods that were used to elucidate or process the structure. For spelling and word order of popular methods, please refer to Method abundancy table and pick those methods appearing more frequently.
|
| BA: |
Biological activity of the compound (free text), including binding information and serological data. If unsure, please indicate whether these data are present in the paper, e.g. serological data.
|
| EI: |
Comma-separated list of enzymes that release or process the structure, including those from other organisms. Please also include enzymes used to process the structure during (chemo-)enzymatic synthesis, if reported by authors. Do NOT include enzymes missing from a strain mutant on the corresponding gene, if absence of this enzyme leads to the described (truncated) structure.
|
| BG: |
Availability of biosynthetic and genetic data in the publication (biochemical data, genetic data)
|
| SY: |
Availability of data on laboratory or industrial synthesis of the compound (chemical, chemical fragmentary, enzymatic, enzymatic in vivo, chemoenzymatic, chemical and enzymatic, synthesis (unknown), fragmentary (unknown), modelling, biosynthesis; leave blank if no synthesis is discussed in the publication).
|
| KW: |
Comma-separated list of keywords from the publication. Greek letters, apostrophs and quotes are supported.
|
| NT: |
Any comments that do not fit other fields (e.g. errors found in the article, reference to revisions, structural elements unencoded in ST1, etc.).
|
| 3D: |
Availability of 3D-structure and conformation data (conformation data, computer modeling, dynamics etc.)
|
| RR: |
Comma-separated list of related CSDB record IDs:
- other structures compared with this structure in the publication,
- fragments of this structure,
- similar structures having only minor differences with this structure,
- structures of the same type from the same organism,
- bigger structures, which this structure is a fragment of,
- other subunits of the same molecule,
- etc.
If multiple structures are published in a review, cross-link them only if they are produced by the same organism or are closely interrelated. If a referenced record is in another domain, precede it with letter B, P or F (depending on the dump it is contained in: bacterial, plant or fungal). If a reference should not be imported, but you still wish to keep it (e.g. referenced record is temporarily excluded from the dump by adding a star), add * afterwards. Example: 1234,B5678,100500*,12345
|
| DB: |
Cross-references to other structural databases: comma-separated list of resource-identifier pairs (resource identifier is before colon, reference is after colon, e.g. CA:123456789). Widespread resource identifiers are CCSD (Carbbank ID),GlycomeDB (GlycomeDB ID), CA (Chemical Abstracts access number), CA-RN (CA substance registry number), US-PT (USA patent number), ProtDB (Protein Databank ID), GenDB (GenBank ID), GTC (Glytoucan ID).
|
| TAX: |
Comma-separated list of cross-references to the NCBI Taxonomy database, according to the order of organisms in the SO field. The number of values should be equal to the number of taxa in SO, excluding taxon remappings in curly brackets. Please specify NCBI TaxID of the organism (strain). If no NCBI TaxID exists for the organism, use NCBI TaxID of the species (or genus if species is missing from the NCBI Taxonomy database) in parentheses. If sp. is used for species and NCBI TaxID of the exact organism is unknown, use NCBI TaxID of the genus. For hybrid organisms, combine two TaxIDs with star (100*101). If a genus is missing from the NCBI Taxonomy database, please use a TaxID of the kingdom and negate it (e.g. -2 for bacteria). A value of -1 means the structure is natural but organism is unknown.
|
| U1: |
Last name of the record submitter (yours).
|
| U2: |
Record submission date (DD.MM.YYYY). Can be rounded to the first day of month.
|
| U3: |
For bacterial papers: RefManID of this paper in the local database of the Carbohydrate Chemistry Lab, Zelinsky Institute. Add PDF if a full-text PDF is present.
|
| U4: |
For bacterial papers: Comma-separated list of RefManIDs of related papers in the local database of the Carbohydrate Chemistry Lab, Zelinsky Institute.
|
| U5: |
If the record was imported from another database (e.g. Carbbank), U5 lists errors found in the original record.
|
| U6: |
Filename (with extension) of the publication full text in CSDB local cache. Please name files using PMID (12345678.pdf) or id_####.pdf (CSDB ID if no PMID is available)
|
Carbohydrate notations are formal languages used to describe or visualize the glycan structure. CSDB native notation is called CSDB Linear and provides a human-readable one-line structure encoding. It is used to formalize search queries, helps error tracking and simplifies data posting and input/output. The comparison of CSDB Linear to other carbohydrate and general chemical notations is shown in the table ( red = worse, green = better). Notations listed in italics are no more supported in modern carbohydrate databases.
CSDB has built-in translators (parsers) from CSDB Linear and GlycoCT and exporters of structures to CSDB Linear, GlycoCT (condensed or XML), Glyde II, LinUCS, WURCS, GLYCAM, SMILES (with deambiguation), structural formula (via SMILES, image), SweetDB (based on IUPAC extended), MOL file (via SMILES, with 3D optimization), PDB file (with residue assignment), SNFG (image).
Structures are retrieved from database or parsed from a notation to an array. Click here to view its description.
To check how a certain structure is parsed, you can use our Structure checker. Scroll to the Check structure syntax section, type in the structure in CSDB Linear encoding, press [Check!] button, and if you see Parsing OK in the output, click on Show/hide parsed data structure.
The structure is parsed into globals:
$errors[] - array of error messages and warnings.
$parsed_structure[] - array for the parsed structure. It has the following keys:
[0]....[n] (numeric keys) => subarray for each residue, key is an index used as id of a residue within the parsed structure
[root_id] => index of the oligomer root residue or the rightmost residue in polymer
[polymer] => true if structure is a repeating unit
[explanations] => array of explanations for residues like Subst1, Sug etc., i.e. what is after '//' in linear code.
keys are names of residues, values are explanations for these residues
Each residue subarray has the following keys:
[stoichiometry] => stoichiometry of the residues in percent (usually 100); '?' if less than 100%, but unknown
[type] => residue type:
'normal' - normal residue
'mva' - monovalent residue (Ac, Me, etc.)
'superclass' - superclass (PEN, HEX, LIP, etc.)
'fuzzy' - a special residue which is a set of alternative variants. Has a special format (see below)
[variant] => true if a residue is one of variants in a set
[variant_set] => if the residue is a variant ('variant' flag is true), this key is the index of the set of variants (fuzzy residue) in which the residue is contained. For other types of residues contains nothing.
[anomeric] => anomeric configuration (a,b,l,x,?)
[absolute] => absolute configuration (D,L,R,S,X,?)
[basename] => basename (Gal, Fuc, 6dTal, etc.)
[ringsize] => ring size (p,f,a,?,'')
[qualifiers] => modifications (N, A, N3N, -ol, etc. in any combination)
[skeleton_size] => number of carbons in the residue
[donates_to] => array with linkage information on the bond in which the residue is a 'donor' (including bonds from variants of a fuzzy set)
usually has one key [0], but can also have [1] if residue is linked dually, or if there are two outgoing linkages (one for inter-repeating subunit, another for its left cap).
Array is empty for a residue at the reducing end.
sub-subkeys are (n = 0 or 1):
[n][residue_id] => id of the acceptor residue. If acceptor is FUZZY, the index of a variant set is provided.
[n][goes_by] => atom by which the residue is linked (usually 1 or 2)
[n][goes_to] => the substituted atom in the acceptor residue
[n][link_type] => link type (glycosidic,amidic,diester,amine,carbon-carbon,other)
[n][external] => true if this acceptor is a part of structure attached to the right residue in sub-repeat (co-exists with regular acceptor, which is the leftmost residue in this sub-repeat); false for all other residues except sub-repeat right end
[n][repeat] => this key is present only for inter-repeat linkage of the rightmost residue in sub-repeat and indicates the number of repeats (e.g. 3, ?, 3-5)
[substituted_by] => array with all donors that are linked to the residue, excluding donors that are variants of a fuzzy set (while fuzzy set itself should be among donors)
Has numeric subkeys, one for each donor. Array is empty for terminal residues.
sub-subkeys are (n = incremental):
[n][donor_id] => id of the donor residue. If donor is FUZZY, all subvariants are listed separately ([0][], [1][], etc).
[n][goes_by] => the atom by which the donor substitutes the residue (usually 1 or 2)
[n][goes_to] => the substituted atom in the residue
[n][link_type] => link type (glycosidic,amidic,diester,amine,carbon-carbon,other)
[n][variant_set] => (optional key) if a donor residue is one of variants, the index of variant set (fuzzy residue) is provided
[n][external] => true if this substituent is a cap attached to the left residue in sub-repeat (co-exists with regular substituent, which is the rightmost residue in this sub-repeat); false for all other residues except sub-repeat left end
[n][repeat] => this key is present only for inter-repeat linkage of the rightmost residue in sub-repeat and indicates the number of repeats (e.g. 3, ?, 3-5)
[repeat] => 'L' for the leftmost residue in the repeating unit, 'R' for the rightmost one, 'RL' if both together, '' for all other residues.
[subrepeats] => subrepeats, which this residue belongs to; array of elements, where keys = subrepeat ID, values = number of repeats (allowed: 3, ?, 3-5). For regular residues (not inside sub-repeating units) this array is empty
[subrepeat_start] = >sub-repeat ID if this residue is leftmost in a subrepeat; false for all other residues
[subrepeat_end] = >sub-repeat ID if this residue is rightmost in a subrepeat; false for all other residues
If the residue type is 'fuzzy' (set of variants), residue subarray has the following keys:
[stoichiometry] => always 100 (non-stoichiometrical substitution is reflected in variants of this variant set)
[type] => 'fuzzy'
[basename] => 'FUZZY'
[fuzzy] => Array describing all variants. Subkeys are:
[logic] => variant combination logic ('OR' or 'XOR')
[n] (numeric keys) => indices of variants (residues), one for each variant
[donates_to] => has only one key: [0]['residue_id']=> id of the acceptor residue
[substituted_by] => (format as above; unsure: is it mandatory?)
[repeat] => (format as above; copy of a value, which should be the same in all variants)
[subrepeats] => (format as above; copy of a value, which should be the same in all variants)
[subrepeat_start] = > (format as above; copy of a value, which should be the same in all variants)
[subrepeat_end] = > (format as above; copy of a value, which should be the same in all variants)
[variant] => (format as above; if this nested fuzzy residue is a variant in another fuzzy residue)
[variant_set] => (format as above; if this nested fuzzy residue is a variant in another fuzzy residue)
Residue subarrays can have the following NMR simulation-related keys:
[lineage] => lineage string (comma-separated substitution positions to track this residue starting from the reducing end or the rightmost residue in a polymer repeat; for reducing end itself it is an empty string)
[NAc] => array of positions (atom numbers) where N-acetyl groups are attached
[props_idx] => index of entry in a database of residue properties
[anomeric_atom] => position of the anomeric atom (usually 1 or 2)
[protonation] => protonation pattern (e.g. 11111203, where character position is the atom number, and character itself indicates the number of protons at this position)
[c13data_idx] => index of entry of this residue in a reference 13C NMR database (for BIOPSEL)
[subspectrum] => 13C NMR data simulated by BIOPSEL (empirical)
[free] => array of chemical shifts of the unsubstitiuted (free) monomer; keys = atom numbers; values (integer) = chemical shifts multiplied by 10
[free_spc_source] => a string label ("reducing" or "linked") indicating the details of obtaining a spectrum of the unsubstitited monomer
[shifts] => array of simulated chemical shifts of the residue in the structure, as if it was on the reducing end (i.e. effects of substituents (bond donors) are applied, but effects on this residue from its bond acceptors are not applied); keys = atom numbers; values (integer) = chemical shifts multiplied by 10
[c1eff] => array of substitution effects to apply to donating atoms by which other residues (bond donors) substitute this residue
[c2eff] => (present if not zero) array of substitution effects to apply to next-to-donating atoms by which other residues (bond donors) substitute this residue
[consistency] => probably:), accuracy level (0-3.5), see explanation at http://csdb.glycoscience.ru/help/nmr.html#emp_hybr
[spectrum_db] => statistically (GODDESS) simulated residue subspectra (1H and 13C)
[C|H] => simulated data; key is a nucleus ('C' or 'H')
[<atom number>] => simulated data for each atom; key is a carbon atom number, according to enumeration scheme in residue service database (http://csdb.glycoscience.ru/database/core/residues.php)
[result] => predicted chemical shift (float, or '?' = could not predict this atom, or '-' = no atom with this number exist, e.g. H1 in Neup)
[trust] => trustworthiness level in percent (float 0-100, '?', '-'), see explanation at http://csdb.glycoscience.ru/help/nmr.html#statistical
[tppm] => expected error in ppm (0.2-5.0 for 13C; 0.02-0.50 for 1H; if out of these bounds, returns a string like "<0.02" or ">0.5"
[weight] => total weight of generalizations applied to structural surrounding to predict this atom
[sd] => standard deviation in a set of chemical shifts used for averaging
[data_used] => sampling size (the number of chemical shifts used for averaging)
[reference] => array of matching chemical shifts (after the latest generalization in the report) in the CSDB; keys = incremental
[<key>] => reference data:
[spectrum_id] => NMR spectrum ID in CSDB
[residue_instance_id] => residue instance ID in CSDB
[atom] => atom number
[report] => report on prediction route = the sequence of structure generalizations (array of strings, one line per generalization)
[0 or 1] => this key is only present for prediction of two protons with the same number; in this case [0] and [1] contain their own [result], [trust], [data_used], [reference], while the averaged data are provided as above. This is only for CH2, where chemical shifts can be different in a pair; for all other cases (C, CH, CH3) there are no [0] and [1].
[color] => residue color in the NMR assignment (3 bytes: 0=>R, 1=>G, 2=>B)
[spectrum_emp] => empirically (BIOPSEL) simulated residue subspectrum (13C); keys are atom numbers
[<atom number>] => simulated data:
[result] => chemical shift (float)
[trust] => trustworthiness level in percent (0-100)
[spectrum_hybr] => result of empirical and statistical simulation hybridization; keys are atom numbers
[<atom number>] => simulated data:
[result] => chemical shift (float)
[dev] => deviation between empirical and statistical simulation, in ppm (float)
[trust] => trustworthiness level in percent (0-100)
[trust] => overall trustworthiness for this residue
[spectrum_c2d] => some other data (?), the format is the same as for [spectrum_hybr]
 XML
XML XML
XML