Taxonomic group: fungi / Basidiomycota
(Phylum: Basidiomycota)
Organ / tissue: fruiting body
NCBI PubMed ID: 39276254Publication DOI: 10.1007/s13659-024-00476-6Journal NLM ID: 101577734Publisher: Heidelberg: Springer
Correspondence: Yang L <yangliu

mail.kib.ac.cn>; Hu JM <hujiangmiao

mail.kib.ac.cn>
Institutions: University of Chinese Academy of Sciences, Beijing, China, Key Laboratory of Phytochemistry and Natural Medicines, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Dictyophora rubrovalvata is a valuable fungus homologous to food and medicine, and its polysaccharide have been gaining increasing attention because of its plentiful activity. However, the structure and activity of its homogeneous polysaccharide have not been studied enough. In this study, two polysaccharides DRP-I and DRP-II were purified from D. rubrovalvata. Their structures were characterized by chemical composition, monosaccharide composition analysis, methylation analysis and nuclear magnetic resonance spectroscopy. The results showed that DRP-I and DRP-II were neutral heteropolysaccharides with molecular weights of 5790 and 12500 Da, respectively, which were composed of mannose, galactose, glucose, xylose and fucose. The main chains were→6)-α-D-Galp-(1→6)-α-D-Galp-(2,1→6)-α-D-Manp-(2,1→6)-α-D-Galp-(1, and branch chains were β-D-Xylp-(1→3)-α-L-Fucp-(1→4)-α-D-Manp-(1→and α-D-Galp-(1→3)-α-D-Galp-(1→. The in vitro immunoactivity assays on dendritic cells showed that DRP-I and DRP-II could up-regulate the expression of IL-10 and IL-6 and inhibit the expression of TNF-α in a concentration-dependent manner. This research indicated that DRP-I and DRP-II possessed immunoactivity by balancing the excessive inflammation, and molecular weight is an important factor affecting immunoactivity
polysaccharides, structure characterization, immunoactivity, Dictyophora rubrovalvata
Structure type: structural motif or average structure ; 5790
Location inside paper: Fig. 3, Table 3
Compound class: polysaccharide
Contained glycoepitopes: IEDB_114701,IEDB_130701,IEDB_134624,IEDB_136045,IEDB_136906,IEDB_137472,IEDB_141794,IEDB_142489,IEDB_144562,IEDB_144983,IEDB_151528,IEDB_152206,IEDB_152214,IEDB_167188,IEDB_174332,IEDB_174333,IEDB_190606,IEDB_983930,SB_163,SB_44,SB_67,SB_7,SB_72,SB_86
Methods: 13C NMR, 1H NMR, NMR-2D, ELISA, acid hydrolysis, biological assays, HPLC, GPC, UV, extraction, acetylation, methylation analysis, reduction, precipitation, phenol-sulfuric acid assay, SEM, derivatization, Sevag method, centrifugation, HPLC-ELSD, FT-IR, proliferation assay, Congo red test, bicinchoninic acid assay
Biological activity: compound could up-regulate the expression of IL-10 and IL-6 and inhibit the expression of TNF-α in a concentration-dependent manner
NCBI Taxonomy refs (TaxIDs): 1464785
Show glycosyltransferases
NMR conditions: in D2O
[as TSV]
13C NMR data:
Linkage Residue C1 C2 C3 C4 C5 C6
6,6,6,6 aDGalp 97.81-97.86 68.25 69.46 68.21 68.78 66.44
6 aDGalp 97.81-97.86 68.25 69.46 68.21 68.78 66.44
6,6,6,2,3 aDManp 98.20 70.37 72.87 66.75 76.22 61.02
6,6,6,2 aDGalp 102.40 69.78 77.57 72.54 68.78 61.02
6,6,6 aDManp 98.30 76.95 70.99 ? ? 66.44
6,6,2,4,3 bDXylp 104.43 73.02 75.65 69.20 65.06
6,6,2,4 aLFucp 101.67 70.37 77.57 73.35 67.19 15.19
6,6,2 aDManp 101.47 68.80 68.25 71.69 ? ?
6,6 aDGalp 97.99 78.05 68.25 67.49 68.25 66.44
aDGalp 97.81-97.86 68.25 69.46 68.21 68.78 66.44
1H NMR data:
Linkage Residue H1 H2 H3 H4 H5 H6
6,6,6,6 aDGalp 5.00-5.02 3.84 4.02 3.89 4.20 3.68-3.91
6 aDGalp 5.00-5.02 3.84 4.02 3.89 4.20 3.68-3.91
6,6,6,2,3 aDManp 5.13 4.11 3.66 3.60 3.38 3.75-3.93
6,6,6,2 aDGalp 5.11 4.08 4.00 3.90 4.16 3.75-3.89
6,6,6 aDManp 5.14 3.94 4.02 ? ? 3.72-3.87
6,6,2,4,3 bDXylp 4.61 3.32 3.45 3.63 3.33-3.96
6,6,2,4 aLFucp 5.09 3.90 4.08 3.77 4.18 1.23
6,6,2 aDManp 5.07 3.79 3.90 3.84 ? ?
6,6 aDGalp 5.06 3.82 4.06 3.97 3.85 3.72-3.87
aDGalp 5.00-5.02 3.84 4.02 3.89 4.20 3.68-3.91
1H/13C HSQC data:
Linkage Residue C1/H1 C2/H2 C3/H3 C4/H4 C5/H5 C6/H6
6,6,6,6 aDGalp 97.81-97.86/5.00-5.02 68.25/3.84 69.46/4.02 68.21/3.89 68.78/4.20 66.44/3.68-3.91
6 aDGalp 97.81-97.86/5.00-5.02 68.25/3.84 69.46/4.02 68.21/3.89 68.78/4.20 66.44/3.68-3.91
6,6,6,2,3 aDManp 98.20/5.13 70.37/4.11 72.87/3.66 66.75/3.60 76.22/3.38 61.02/3.75-3.93
6,6,6,2 aDGalp 102.40/5.11 69.78/4.08 77.57/4.00 72.54/3.90 68.78/4.16 61.02/3.75-3.89
6,6,6 aDManp 98.30/5.14 76.95/3.94 70.99/4.02 ?/? ?/? 66.44/3.72-3.87
6,6,2,4,3 bDXylp 104.43/4.61 73.02/3.32 75.65/3.45 69.20/3.63 65.06/3.33-3.96
6,6,2,4 aLFucp 101.67/5.09 70.37/3.90 77.57/4.08 73.35/3.77 67.19/4.18 15.19/1.23
6,6,2 aDManp 101.47/5.07 68.80/3.79 68.25/3.90 71.69/3.84 ?/? ?/?
6,6 aDGalp 97.99/5.06 78.05/3.82 68.25/4.06 67.49/3.97 68.25/3.85 66.44/3.72-3.87
aDGalp 97.81-97.86/5.00-5.02 68.25/3.84 69.46/4.02 68.21/3.89 68.78/4.20 66.44/3.68-3.91
1H NMR data:
| Linkage | Residue | H1 | H2 | H3 | H4 | H5 | H6 |
|---|
| 6,6,6,6 | aDGalp | 5.00
5.02 | 3.84 | 4.02 | 3.89 | 4.20 | 3.68
3.91 |
| 6 | aDGalp | 5.00
5.02 | 3.84 | 4.02 | 3.89 | 4.20 | 3.68
3.91 |
| 6,6,6,2,3 | aDManp | 5.13 | 4.11 | 3.66 | 3.60 | 3.38 | 3.75
3.93 |
| 6,6,6,2 | aDGalp | 5.11 | 4.08 | 4.00 | 3.90 | 4.16 | 3.75
3.89 |
| 6,6,6 | aDManp | 5.14 | 3.94 | 4.02 | ? | ? | 3.72
3.87 |
| 6,6,2,4,3 | bDXylp | 4.61 | 3.32 | 3.45 | 3.63 | 3.33
3.96 | |
| 6,6,2,4 | aLFucp | 5.09 | 3.90 | 4.08 | 3.77 | 4.18 | 1.23 |
| 6,6,2 | aDManp | 5.07 | 3.79 | 3.90 | 3.84 | ? | ? |
| 6,6 | aDGalp | 5.06 | 3.82 | 4.06 | 3.97 | 3.85 | 3.72
3.87 |
| | aDGalp | 5.00
5.02 | 3.84 | 4.02 | 3.89 | 4.20 | 3.68
3.91 |
|
13C NMR data:
| Linkage | Residue | C1 | C2 | C3 | C4 | C5 | C6 |
|---|
| 6,6,6,6 | aDGalp | 97.81
97.86 | 68.25 | 69.46 | 68.21 | 68.78 | 66.44 |
| 6 | aDGalp | 97.81
97.86 | 68.25 | 69.46 | 68.21 | 68.78 | 66.44 |
| 6,6,6,2,3 | aDManp | 98.20 | 70.37 | 72.87 | 66.75 | 76.22 | 61.02 |
| 6,6,6,2 | aDGalp | 102.40 | 69.78 | 77.57 | 72.54 | 68.78 | 61.02 |
| 6,6,6 | aDManp | 98.30 | 76.95 | 70.99 | ? | ? | 66.44 |
| 6,6,2,4,3 | bDXylp | 104.43 | 73.02 | 75.65 | 69.20 | 65.06 | |
| 6,6,2,4 | aLFucp | 101.67 | 70.37 | 77.57 | 73.35 | 67.19 | 15.19 |
| 6,6,2 | aDManp | 101.47 | 68.80 | 68.25 | 71.69 | ? | ? |
| 6,6 | aDGalp | 97.99 | 78.05 | 68.25 | 67.49 | 68.25 | 66.44 |
| | aDGalp | 97.81
97.86 | 68.25 | 69.46 | 68.21 | 68.78 | 66.44 |
|
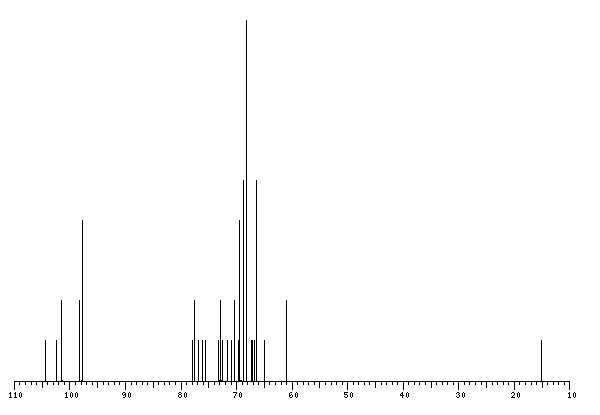
The spectrum also has 4 signals at unknown positions (not plotted). |
There is only one chemically distinct structure:
 report error
report error Found 1 record.
Displayed record 1
Found 1 record.
Displayed record 1
 report error
report error
 mail.kib.ac.cn>; Hu JM <hujiangmiao
mail.kib.ac.cn>; Hu JM <hujiangmiao mail.kib.ac.cn>
mail.kib.ac.cn>