| Enzyme / Gene |
Activity |
Object |
References |
|---|
Enzyme name: LpxL
UniProt ID: P0ACV0 
Gene name: lpxL
Gene GenBank ID: 946216*  | Synthesized dimer: Lau(1-3)R3HOMyr
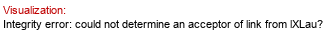lR3HOMyr?)
Status: indirect evidence in vivo
Confirmation methods: mutation (deletion)
ID: 2317 Notes: LpxL particilates in lauroylation of lipid A; its deletion led to the addition of
P-EtN or L-Ara4N to the pentaacylated lipid A, as well as of P-EtN to the tetraacylated
lipid A, at its phosphate groups.
| Organism (ID 23): Escherichia coli K12
Full structure (ID 1795):
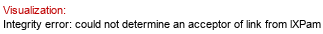lR3HOMyr(1-2)[lXLau(1-3)lR3HOMyr(1-2)[P-4),lXMyr(1-3)lR3HOMyr(1-3)]bDGlcpN(1-6),lR3HOMyr(1-3)]aDGlcpN(1-P&highlight=4~3~lXLau)
CSDB ID(s): 681, 810, 811, 2492, 8831, 12409, 31184, 31285, 31328, 32011Molecule role: lipid A | Klein et al. 2009
DOI: 10.1074/jbc.M900490200
|
#GTR ID Enzyme name Enzyme group Enzyme Genbank ID Derived ID mark Enzyme Uniprot ID Derived ID mark CAZY family Gene name Gene Genbank ID Derived ID mark Gene cluster Full structure CSDB structure ID Side product CSDB structure ID Full structure Full structure is fragment (0|1) Synthesized bond Synthesized bond location (from root ~ subst position) Donor structure ID Substrate structure Confirmation status Methods used Notes Molecule role Molecule is a model of role (0|1) Molecule is a precursor of role (0|1) Main CSDB record ID Additional CSDB record IDs CSDB organism IDs Organism names Organs and tissues References (;-separated)
2317 LpxL P0ACV0 lpxL 946216 * 1795 lXPam(1-3)lR3HOMyr(1-2)[lXLau(1-3)lR3HOMyr(1-2)[P-4),lXMyr(1-3)lR3HOMyr(1-3)]bDGlcpN(1-6),lR3HOMyr(1-3)]aDGlcpN(1-P 0 lXLau?(1-3)lR3HOMyr? 4~3 indirect mutation (deletion) LpxL particilates in lauroylation of lipid A; its deletion led to the addition of P-EtN or L-Ara4N to the pentaacylated lipid A, as well as of P-EtN to the tetraacylated lipid A, at its phosphate groups. lipid A 0 0 681,810,811,2492,8831,12409,31184,31285,31328,32011 23 Escherichia coli K12 Klein et al. 2009 (doi: 10.1074/jbc.M900490200)

